In the RAMIS project, we had to characterize the DNA organization (i.e., chromatin texture) in cell nuclei. More precisely, the ”quantity of DNA” is represented by the intensity of the pixel (i.e., the higher the pixel gray level, the higher quantity of DNA). As we observe from the example bellow, the distribution of DNA is not stationary and, for some classes, it is usually closer to the center of the nuclei.

Then, in order to characterize such radial textures, we propose a descriptor named gray level Distance (to border) Zone Matrix (DZM), denoted GDf (dn, gm). The new statistical matrix yields the number of zones of intensity gm at a distance of dn from the border of support space Ec (shortest Euclidean distance).
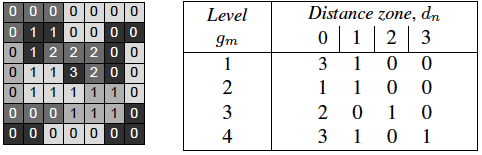
You can see in this paper that this method has become a standard in radiomics.
It is obvious how to generalize this matrix to construct a Multiple gray level Distance (to border) Zone Matrix.
On this page is a software to fill the matrix and compute the features.
Do not hesitate to contact me if you need some explanations.
Related publication:
- 1 - Advanced Statistical Matrices for Texture Characterization: Application to DNA Chromatin and Microtubule Netwok Classification, IEEE ICIP 2012.
- 2 - Advanced Statistical Matrices for Texture Characterization: Application to Cell Classification, IEEE Transaction on BioMedical Engineering 2014.